The clinical outcomes of pediatric acute myeloid leukemia (AML) have shown improvement with combination therapies; however, relapse remains a significant concern due to the resistance of leukemia cells to conventional treatments. In some cases, epigenetic heterogeneity within leukemias can contribute to therapy resistance. For example, leukemia stem cells (LSCs) have been shown maintain AML, both before and after therapy. This study aims to identify LSCs within a mouse model of high-risk, NUP98-rearranged pediatric AML, and to identify mechanisms that sustain this population.
To identify and isolate a LSC subpopulation, we characterized murine NUP98-HOXD13/Flt3-ITD-driven AML in mice by performing cellular indexing of transcriptomes and epitopes (CITE-seq) analysis at two stages: pre-AML and fully transformed AML. Uniform Manifold Approximation and Projection (UMAP) revealed a highly reproducible pattern of heterogeneity within each AML specimen. Each AML had a small percentage of cells that resembled normal and pre-leukemic multipotent progenitors (MPPs), as well as additional populations with B-cell or myeloid gene expression. Pseudotime analysis indicated a trajectory from MPPs to mature cell-like lineages, implicating the MPP-like population as a potential LSC. This population expressed type I interferon (IFN-1) target genes more highly than non-LSC AML cells and pre-AML cells. This finding is significant because we previously identified IFN-1 as an effector of Flt3-ITD/NUP98-HOXD13 synergy during AML initiation, and in human NUP98-rearranged AML (PMID: 36395068).
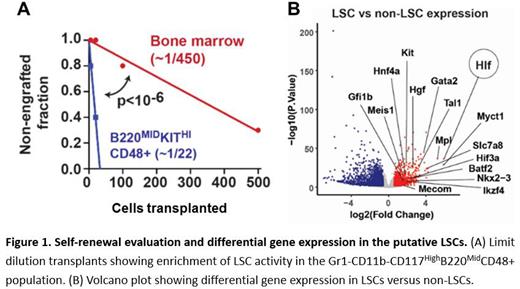
Differential gene expression analysis also identified several established self-renewal genes, including Hlf, Myct1, Mpl, Slc7a8, and Gata2, as highly expressed in putative LSCs relative to non-LSCs (figure B). This suggests that AML LSCs superimpose inflammatory signaling programs, particularly IFN-1, and core stem cell self-renewal programs to maintain a malignant state. The data raise the question of how IFN-1 shapes LSC identity.
We next sought to isolate and validate the putative LSC population. CITE-seq feature barcode data indicated high expression of CD117 and CD48, medium expression of B220, and low expression of Gr1 and CD11B in putative LSCs. We isolated LSCs based on these markers (Gr1-CD11b-B220 MidCD117 highCD48+). Several non-LSC populations were isolated for comparison (e.g., Gr1+CD11B+ and Gr1-CD11B-B220 highCD117 Mid populations. Cytospin observation revealed no morphological differences among the three populations, but cell culture assays showed that only the Gr1-CD11b-B220 MidCD117 highCD48+ cells exhibited proliferative capacity after multiple passages. Limit dilution transplant assays showed that the Gr1-CD11b-B220 MidCD117 highCD48+ population was greatly enriched for LSC function relative to whole bone marrow (figure A).
Altogether, we have shown that we can prospectively isolate a LSC population from NUP98-HOXD13/Flt3-ITD AML. The LSC population is highly reproducible across specimens and it demonstrates high degrees of IFN-1 activity. Further work will establish the role of IFN-1/self-renewal gene interactions in LSC maintenance.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal